This is the Github repo of CODA-19: Using a Non-Expert Crowd to Annotate Research Aspects on 10,000+ Abstracts in the COVID-19 Open Research Dataset.
CODA-19 is a human-annotated dataset that denotes the Background, Purpose, Method, Finding/Contribution, and Other for 10,966 English abstracts in the COVID-19 Open Research Dataset.
CODA-19 was created by 248 crowd workers from Amazon Mechanical Turk collectively within ten days. Each abstract was annotated by nine different workers, and the final labels were obtained by majority voting.
CODA-19's labels have an accuracy of 82% and an inter-annotator agreement (Cohen's kappa) of 0.74 when compared against expert labels on 129 abstracts.
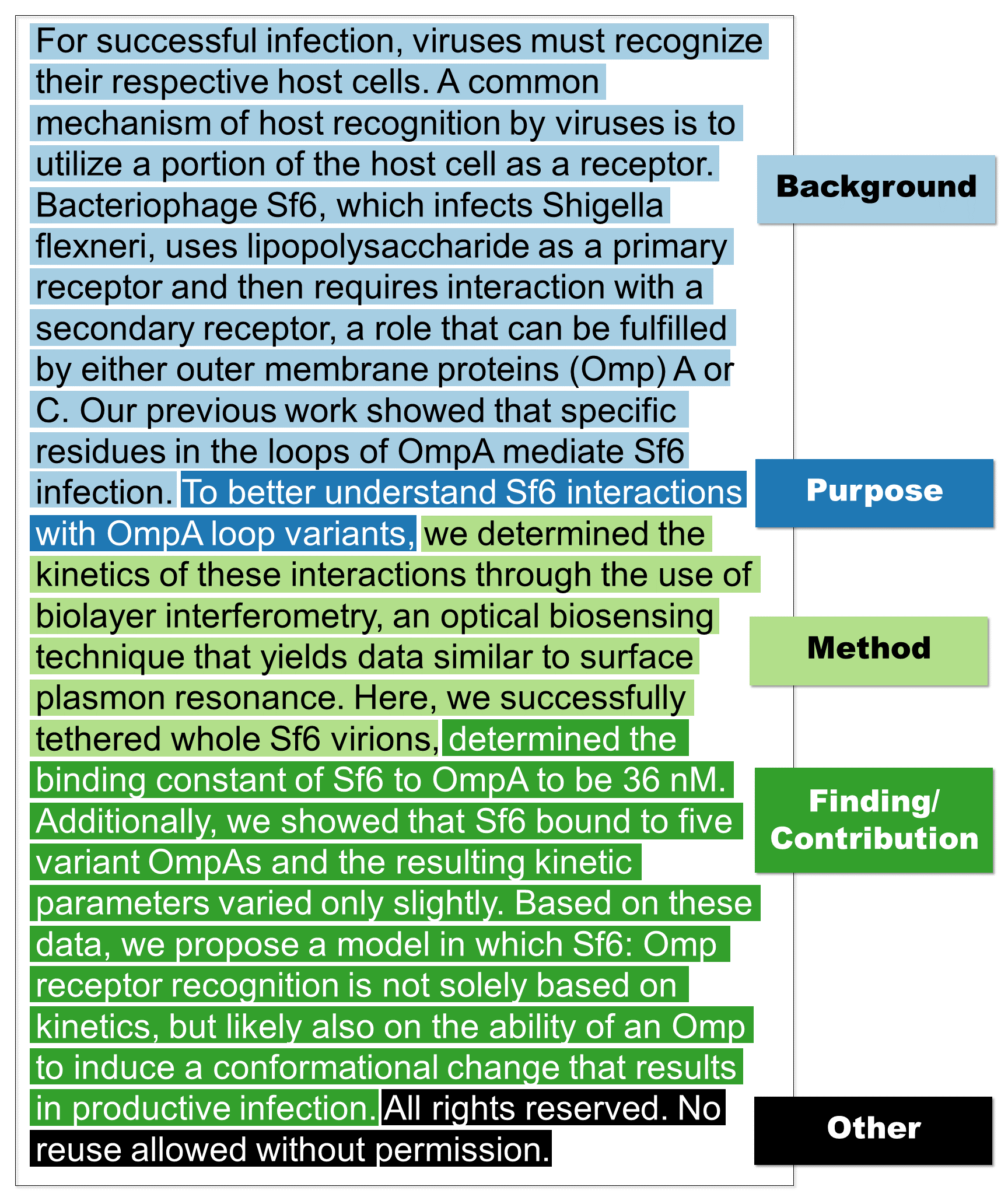
The following is an actual abstract (you can see the paper here) annotated by crowd workers in CODA-19.
You can download the worker interface we used to construct CODA-19 from here: Download CODA-19 Interfaces (~50MB).
These are the interfaces of the last batch of HITs we posted to MTurk when constructing CODA-19. You can download them and simply open any HTML file on your local laptop.
We iteratively improved the instruction and worker interface for each batch. These are the last batch, so the instructions are the most complete.
Yes, we made our baseline classifiers avalible. Please go to the classification folder of this repo.
This work was developed upon the long history of research on understanding scientific papers at scale. In short, the rapid acceleration in new coronavirus literature makes it hard to keep up with. So we highlighted the papers with its Background, Purpose, Method, Finding/Contribution, and Other. People can use these data to build an automated annotator to label the remaining papers in CORD-19 and, more importantly, future papers. This type of annotation can also be useful for various BioNLP tasks.
We used Stanford CoreNLP to tokenize and segment sentences for all the abstracts in CORD-19. We further used comma (,), semicolon (;), and period (.) to split each sentence into shorter fragments, where a fragment has no less than six tokens (including punctuation marks) and has no orphan parentheses. As of April 15, 2020, 29,306 article in CORD-19 had a non-empty abstract.
We filtered out the 538 (1.84%) abstracts with only one sentence and the 145 (0.49%) abstracts that had more than 1,200 tokens. We randomly selected 11,000 abstracts from the remaining data for annotation.
During the annotation process, workers informed us that a few articles were not in English. We identified them automatically using langdetect and excluded them. The released version of CODA-19 has totally 10,966 abstracts.
├── human_label # human labels folder
│ ├── test # test set, containing 1,000 abstracts
│ │ └── expert # expert labels folder
│ │ ├── biomedical_expert # expert labels from Bio expert
│ │ ├── computer_science_expert # expert labels from CS expert
│ │ ├── biomedical_expert-eval.csv # crowd labels evaluated against Bio expert's labels
│ │ └── computer_science_expert-eval.csv # crowd labels evaluated against CS expert's labels
│ ├── dev # dev set, containing 1,000 abstracts
│ ├── train # training set, containing 8,965 abstracts
│ └── coda_metadata.csv # metadata for CODA-19
└── machine_label # empty folder (desgined for future automatic labels)
{
"paper_id": the paper ID in CORD-19,
"metadata": {
"title": the title of the paper,
"coda_data_split": test/dev/train in CODA-19,
"coda_paper_id": numeric id (starting from 1) in CODA-19,
"coda_has_expert_labels": if this paper comes with expert labels in CODA-19,
"subset": the subset (custom_license/biorxiv_medrxiv/comm_use_subset/noncomm_use_subset) in CORD-19
},
"abstract": [
{
"original_text": the tokenized text of the paragraph 1,
"sentences": [
[
{
"segment_text": the tokenized text of the text segment 1 in sentence 1 in paragraph 1,
"crowd_label": the label derived (e.g., majority vote) from a set of crowd labels
},
{
"segment_text": the tokenized text of the text segment 2 in sentence 1 in paragraph 1,
...
},
...
],
[
{
"segment_text": the tokenized text of the text segment 1 in sentence 2 in paragraph 1,
...
},
...
],
...
]
}
{
"original_text": the tokenized text of the paragraph 2,
"sentences": [
...
]
},
...
],
"abstract_stats": {
"paragraph_num": the total number of paragraphs in this abstract,
"sentence_num": the total number of sentences in this abstract,
"segment_num": the total number of text segments in this abstract,
"token_num": the total number of token in this abstract
}
}
We worked with a biomedical expert and a computer scientist to assess the label quality. Both experts respectively annotated the same 129 abstracts randomly selected from CODA-19. The inter-annotator agreement (Cohen's kappa) between two expert is 0.788. Table 2 shows the aggregated crowd label's accuracy, along with the precision, recall, and F1-score of each class. CODA-19's labels have an accuracy of 0.82 and a kappa of 0.74, when compared against two experts' labels. It is noteworthy that when we compared labels between two experts, the accuracy (0.850) and kappa (0.788) were only slightly higher.
Annotating one abstract costs $3.2 on average with our setup. This cost includes the payments for workers and the 20% fee charged by mturk.
Our current budget allowed us to annotate ~11,000 abstracts. If you are interested in funding this annotation effort, please contact Kenneth at txh710@psu.edu).
@inproceedings{huang2020coda,
title={CODA-19: Using a Non-Expert Crowd to Annotate Research Aspects on 10,000+ Abstracts in the COVID-19 Open Research Dataset},
author={Huang, Ting-Hao Kenneth and Huang, Chieh-Yang and Ding, Chien-Kuang Cornelia and Hsu, Yen-Chia and Giles, C Lee},
year={2020},
booktitle={Proceedings of the 1st Workshop on {NLP} for {COVID-19} at {ACL 2020}},
month = jul,
volume = 1,
address = "Online",
publisher = "Association for Computational Linguistics",
}
This project is supported by the Huck Institutes of the Life Sciences' Coronavirus Research Seed Fund (CRSF) at Penn State University and the College of IST COVID-19 Seed Fund at Penn State University. We thank the crowd workers for participating in this project and providing useful feedback. We thank VoiceBunny Inc. for granting a 60% discount for the voiceover for the worker tutorial video to support projects relevant to COVID-19. We also thank Tiffany Knearem, Shih-Hong (Alan) Huang, Joseph Chee Chang, and Frank Ritter for the great discussion and useful feedback.
-
We presented our work at the NLP COVID Workshop @ ACL 2020. July 09, 2020.
-
IST seed grants support tech projects related to COVID-19. May 13, 2020. PSU News.
-
Human and AI annotations aim to improve scholarly results in COVID-19 searches. April 17th, 2020. Jordan Ford. PSU News.
-
Seed grants jump-start 47 interdisciplinary teams to conduct COVID-19 research . April 14, 2020. Sara LaJeunesse. PSU News.